Unbinding ligands from the 5-HT3 receptor
Unlike the G-protein coupled members of the serotonin receptor family, the 5-HT3 receptor (5-HT3R) is unique as it is a ligand-gated ion channel (LGIC). It belongs to the Cys-loop family of LGIC that also includes nACh, glycine and GABA A receptors. Similar to all Cys-loop members, the 5-HT3R is a pentameric arrangement of subunits with distinct extracellular and transmembrane domains. Evidence from the 2.1 Å structure of the closely related AChBP has revealed that ligand binding occurs at the interface of two adjacent subunits (Celie et al., 2004; Unwin, 2005) . To understand the process of ligand unbinding in 5-HT3R, we have used homology models of the 5-HT3R extracellular domain based upon the structure of acetylcholine binding protein (AChBP) and performed molecular dynamic (MD) simulations. We have previously docked granisetron and 5-HT into a homology model using AUTODOCK (Reeves et al., 2003; Thompson et al., 2005) . In the simulations we used the orientation of ligands that were best supported by experimental data as the starting point. Simulations were performed using the mutual repulsion method where the distance between the centres of mass of the ligand and the receptor were incrementally increased during the simulation, which prevents the prior imposition of a specific unbinding trajectory (Chau, 2001) . Four simulations were performed for each ligand using variations in tethering and velocity rescaling.
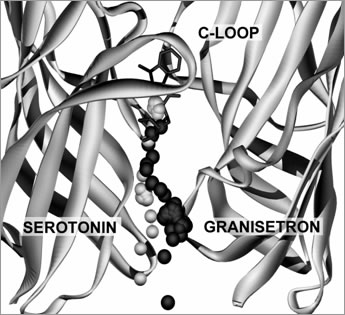
Figure 1. Typical unbinding trajectories for 5-HT and granisetron from the 5-HT3R binding site. Inset shows the location of the larger image within a 5-HT3R dimer. The MD simulations revealed that ligands leave the 5-HT3R binding site along a tunnel that is formed at the interface of two adjacent subunits. The pathway extends down and out from the base of the binding site for 8 Å and widens to form an entrance at a location 20 Å above the membrane. All the trajectories for granisetron and 5-HT were similar (figure 1), showing that tethering and velocity rescaling had no significant effect. Our results show that both 5-HT and granisetron leave the ligand binding site along a trajectory that is formed by a tunnel of amino acids that extends towards the outer surface of the receptor. Celie, P. H. et al. (2004) Neuron 41 , 907-914. |