Computational modelling to aid drug discovery in GPCRs The mechanism by which many GPCR ligands bring about efficacious and off-target effects is poorly understood and this can hinder the drug development process1. Computational models can be used to better understand drug-receptor interactions and downstream effects2. These virtual experiments can provide insight into data and motivate new experiments. We use a system of ordinary differential equations (ODEs) to quantitatively model important reactions in a GPCR signalling pathway. Generating quantitative models of mammalian signal transduction can be difficult due to competing signalling components. Therefore we use a simple system with a small number of quantifiable components. The yeast pheromone response is a useful tool with which to analyse individual GPCR signalling pathways and the simplicity of the yeast system enables the development of robust generic models. We have used a modified yeast system3 to develop a quantitative ODE model of human adenosine A1 receptor (A1R) pharmacology. 
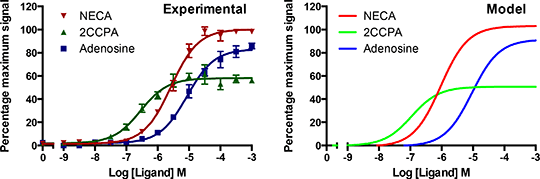
Figure 1. The GPA1/ Gái3 yeast strain expressing the A1R was stimulated with the shown ligands and the data analysed using the operational model for receptor agonism2. Parameter estimation techniques were applied to a modified ODE model4 to replicate the experimental data. The A1R has been functionally expressed in yeast strains representing the inhibitory G proteins Gαo, Gαi1, Gαi3 and Gαz. We have stimulated these strains with the agonists NECA, 2CCPA and adenosine (log Ka = -4.82, -3.41 and -5.84, log τ = 1.12, 1.55 and 0.35 respectively for the GPA1/Gαi3 strain, Figure 1). An established ODE model of the yeast GPCR signalling pathway4 has been modified to suit the A1R in these strains. Parameter estimation techniques were used to fit the model to the experimental data (Figure 1, r2 = 0.99). Using this model we have predicted several rate constants for the interaction of NECA, 2CCPA and adenosine with the A1R and downstream effects on the G protein cycle. Our predictions have been based on experimental end-point data. The challenge is to validate our model predictions by fitting the model to concentration-response time-course data. This can be done using fluorescent biosensors and A1R specific fluorescent ligands. Using the A1R we seek to develop a generic quantitative model of GPCR signalling in yeast. We then envisage applying our model predictions to mammalian systems to augment the drug discovery process. References 1 Langemeijer EV et al, Purinergic signalling, 9:91, 2012. 2 Black JW & Leff, P, Proc. R . Soc. Lond. B. 220:141, 1983. 3 Brown AJ et al, Yeast, 16:11, 2000. 4 Smith B et al, Cellular signalling, 21:1151. |