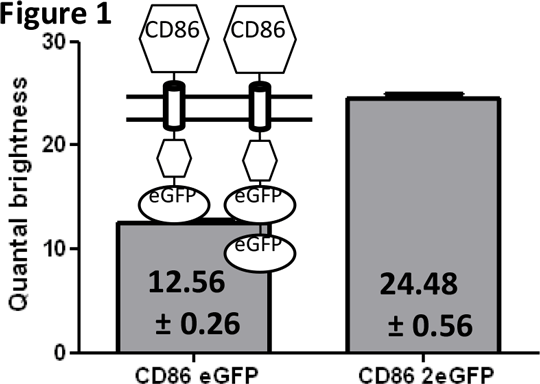
Using Spatial Intensity Distribution Analysis to determine the oligomeric structure of the Roundabout 1 receptor. Novel approaches are needed explore the basal oligomeric assembly of transmembrane proteins and how this may be altered in response to extracellular ligands and other cellular challenges. Spatial Intensity Distribution Analysis (SpIDA) (1) is a methodology for determining oligomeric size and the relative abundance of different oligomeric species, based upon the analysis of laser scanning confocal images of cells expressing the protein of interest fused to a fluorescent label. The software with which the analysis is carried out measures the level of fluctuation of fluorescent signal within each pixel and uses this to calculate a value for the quantal brightness of the particular fluorescent label. Thus a doubling of the quantal brightness suggests the presence of a dimer. To validate the application of SpIDA we employed two systems. In the first the quantal brightness values of constructs consisting of the (known monomeric) cluster of differentiation protein CD86 when fused to either one or two copies of enhanced Green Fluorescent Protein (eGFP) was assessed following stable expression of these constructs in HEK293 cells (Figure 1). The 2x eGFP containing construct was found to have twice the quantal brightness of the 1x eGFP form. (CD86 eGFP n=360, CD86 2xeGFP n=240, values are mean+/- SEM).
 
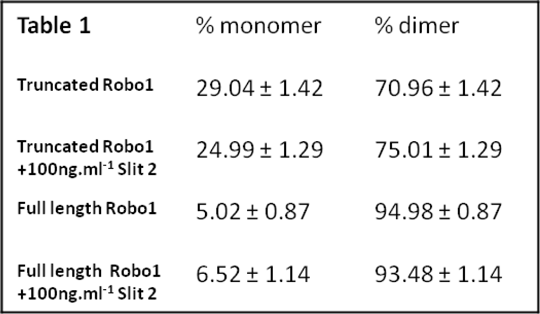
To assess the capacity of SpIDA to monitor the kinetics of ligand-induced transition from monomer to dimer and ligand-concentration dependence an Epidermal Growth Factor (EGF) Receptor-eGFP construct was expressed stably. Addition of EGF resulted in rapid changes in quantal brightness, consistent with transition of the receptor from a predominantly monomeric state to being largely a dimer and this was achieved with an EC50 value of 2.72 ± 0.87nM. We extended such studies to the single transmembrane receptor Roundabout 1 (Robo 1) (2). Robo receptors and their ligands (Slits) are involved in neuronal development and possibly angiogenesis and pancreatic cancer. Little is known of the quaternary structure of Robo 1. Robo 1 was labelled with eGFP as both full length and C-terminally truncated forms, these were stably expressed and confocal images of the baso-lateral membrane collected. SpiDA analysis showed that Robo1 was largely a dimer in the basal state and whilst Slit 2 treatment had no effect upon the level of dimerisation (Table 1), C-terminal truncation of Robo1 reduced the extent of dimerisation, suggesting a role for this region in dimer formation and/or stabilisation. (Table 1, n=250, values are mean +/- SEM). (1) Barbeau A et al Methods Enzymol 522:109 2013 (2) Zakrys L et al (Submitted)
|