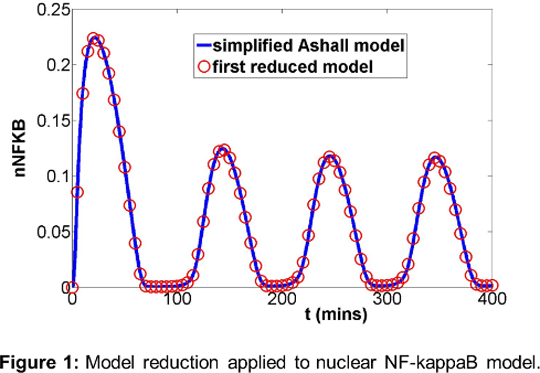
Computational analysis in cell signalling: crosstalk, function and minimal model development for NF-kappaB dynamics Nuclear Factor kappa B (NF-kappaB) is a key transcriptional regulator of cellular responses to stress and infection. Much theoretical and experimental work has been carried out in recent years towards understanding the dynamics of the NF-kappaB system. Key discoveries include oscillations single-cell nuclear NF-kappaB oscillations due to nuclear-cytoplasmic translocations1, dependence of translocation amplitude on pulsatile stimulus frequency, and the functional role of oscillations in controlling gene expression pattern2. Mathematical modelling and simulation are proven and powerful tools for analysis of negative feedback in the system which underlies these oscillations. We review systems and computational approaches underpinning these advances, and present analyses of a new model reduction algorithm applied to NF-kappaB signalling, and crosstalk models for NF-kappaB and other pathways. Model reduction in cell signalling: Mathematical models developed for analysing signalling systems are often complex, with large numbers of variables and parameters. A reduction in order of a model may aid mathematical analysis, and give a representation of key critical components and sub-structures of the original system that were previously unclear. A new, systematic model reduction method based on “speed coefficients” has been developed which reduces the dimension and complexity of model systems, identifying core reactions and variables3. Coupled oscillator behaviour: NF-kappaB signalling dynamics have been seen to vary during the cell cycle4. In particular, dynamic behaviour of the NF-kappaB system depends on cell cycle phase during which the stimulus TNFalpha is added. Experimental and mathematical investigations into the interaction between NF-kappaB and cell cycle regulatory signalling suggests an important interaction behind these effects. Key results: Our new model reduction technique has been applied to 10th order2 and 6th order5 NF-kappaB models, resulting in 4th order models which preserve limit cycle behaviour and bifurcation structure (see Figure 1).

Physical interaction between fluorescently labelled E2F-1 (a cell cycle regulator) and NF-kappaB in SK-N-AS cells was evident using live cell imaging techniques, and a new coupled oscillator model (based on 2,6) which incorporates this detail recapitulates the observed effect of cell cycle on NF-kappaB while also predicting an observed reciprocal effect of NF-kappaB signalling on cell cycle duration. Conclusions: Our new model reduction method appears to have the potential to significantly simplify cell signalling models while preserving complex output features. Furthermore, the elucidation of the control and function of NF-kappaB dynamics continues to require the use of mathematical modelling of the NF-kappaB system and its interaction with other core cellular processes. 1.Nelson et al.,Science, 306:704-708, 2004 2.Ashall et al., Science, 324: 242-246, 2009 3.S. West et al. SUBMITTED. 4. Ankers et al. SUBMITTED. 5. Krishna et al., PNAS, 103(29):10840-1084, 2006. 6. Gerard & Goldbeter, R Soc J Interface Focus, 1:24-35, 2011
|