Applying novel computational techniques to mathematically determine kinetic parameters of GPCR signal transduction. The development of new efficacious drugs is a major challenge to the pharmaceutical industry. Despite continued improvements in production, costs continue to increase, while the number of approved drugs declines. This has been particularly evident in the development of drugs aimed at G protein-coupled receptors (GPCRs), a leading pharmaceutical target. Consequently, new approaches are required. Systems pharmacology (SP) is an emerging discipline combining mathematical and computational techniques to provide a more holistic view of pharmacology and unlike other modelling approaches SP aims to only model what can be measured experimentally. GPCR signalling controls eukaryotic physiology. It is complex with many interlinking dynamic processes occurring over multiple time scales, ranging from subsecond to hours. Novel SP approaches now offer the potential to provide powerful tools for examining GPCR pathways and motivate new experiments. However, the rigorous testing of dynamic GPCR models is only in its infancy due to the lack of high quality, reproducible, uncluttered, dose-response time-data. Here we describe our efforts to develop and apply SP approaches to quantitatively model dose-dependent time-course data derived from a simple GPCR signal transduction cascade.

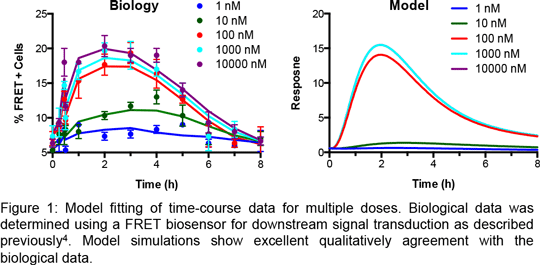
Yeast contains a single combination of GPCR and G protein thereby providing a simplified system in which to study and model receptor pharmacology1. Recently, we have described the production of two ordinary differential equation models that qualitatively simulates dose-response data2,3. Here we extend these models to simulate, for the first time, time-course data for multiple doses derived from the use of intracellular FRET biosensors4 for activation of the downstream signal transduction cascade (Figure 1). We apply computational parameter estimation methods to determine previously un-measurable parameters within our models. Utilising an iterative process of combining experimental observations with parameter estimation we have been able to refine our model to provide excellent qualitative agreement with our biological observations. These observations have extended our knowledge of the modelling process and should be directly transferable to the more complicated systems contained in higher eukaryotes. 1. Ladds et al., (2005) Trends Biotechnol 23: 367-373. 2. Smith et al., (2009) Cell Signal 21: 1151-1160. 3. Croft et al., (2013) J Biol Chem 288: 27327-27342. 4. Weston et al., (2013) PLoS One 8: e77487.
|