A potential role for N-Glycosylation of the orphan receptor GPR61 inmembrane expression
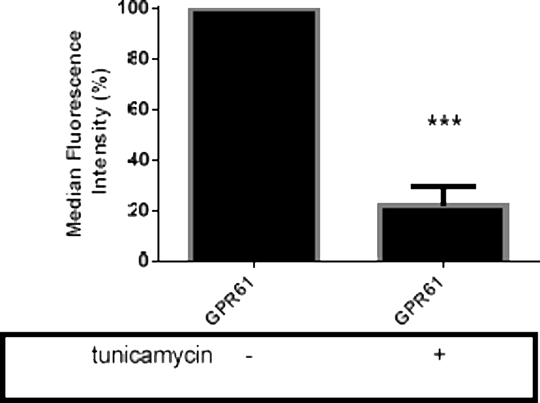
The G-protein coupled receptor 61 (GPR61, BALGR) is an orphanreceptor that is highly expressed in the brain. It has some structural similarity to histamine H2 and 5-HT6 receptors and may be constitutively active(1, 2).The cellular function of GPR61 is unknown, although knock-out studies suggest a role in the regulation of food intake and body weight(3).The primary structure of GPR61 reveals a consensus N-glycosylation site (N12; 1) and, in the present study, we investigate the presence and the role of N-glycosylation HEK293 and COS-1 cells were transiently transfected with a pCMV6vector that included coding for N-terminally His-myc tagged human GPR61. Transfected cells were cultured in the absence or presence of the N-glycosylation inhibitor tunicamycin (1.0 µg/ml) for 48 hours. Harvested cells were assessed for myc-immunore activity by SDS-PAGE/western blotting and cell surface flow cytometry. While untreated cells generated a myc-immunoreactive product around 53 kDa, consistent with the estimated size of the myc-tagged GPR61, the presence of tunicamycin consistently reduced the apparent molecular weight of the resulting myc-immunoreactivity by 1-2 kDa (three independent experiments; 1). Analysis of extracellular myc-immunoreactivity by flow cytometry demonstrated tunicamycin reduced expression of GPR61 at the cell surface (Fig. 1).
Figure 1. Extracellular myc-immunoreactivityexpressed by COS-1 cells transiently transfected withpCMV6 encoding themyc-tagged GPR61 receptor. Cells were cultured in the absence (-) or presence (+) of the N-glycosylation inhibitor, tunicamycin. Data represents mean ± SEM, n=3. Comparison of myc-immunoreactive levels with untreated cells,*** P<0.001 (unpaired t-test)
In summary, the present study suggests GPR61 is subject to N-glycosylation, which may facilitate membrane expression of the heterologously expressed receptor. (1) Lee et al. (2001).Brain Res Mol Brain Res 86: 13–22. (2) Takeda et al. (2003). Life Sci 74: 367–377. (3) Nambu et al. (2011). Life Sci 89: 765–772.
|