Supporting projects, large and small: optimising in vitro pharmacology data delivery for therapeutic projects
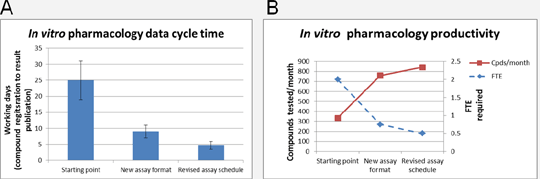
The delivery of new drugs to patients is hampered by a continuing high attrition rate in drug discovery projects (1). Using more efficient drug discovery processes and disease-relevant assay systems should improve the timing and quality of decision-making, enabling the progression of only those projects with the best chance of success (2). Delivery of in vitro pharmacology data to therapeutic projects is a critical element of the optimisation of compounds towards drug-like molecules. Based on these data, decisions are made whether to continue or stop specific areas of medicinal chemistry or antibody optimisation. To make the best decisions, data need to be of sufficient quality and relevance to target pathobiology. Further, the data are provided to projects quickly and efficiently, minimising time spent on unproductive areas of optimisation. There are data available showing improvements in medicinal chemistry (2) and in vitro pharmacology (3) processes, but data characterising refinements across the entire synthesis and test cycle, including measures of quality, are lacking. We have sought to optimise the pharmacological characterisation of small molecules and antibodies in therapeutic projects. As a test case, we mapped the screening cycle for the primary assay carried out for a small molecule therapeutic project by the Primary Pharmacology Group at UCB, Slough, UK. From this, several changes were made to reduce the cycle time between compound registration and result publication, including; 1, changing the format of the primary assay; 2, introduction of an informatics tool, Screen, providing an automated compound ordering solution; 3, implementation of data handling business rules; and, 4, optimisation of the compound delivery and assay schedule. Together, these changes resulted in a reduction in cycle time from >20 working days to < 5 working days (Fig 1A). The improvement in cycle time was accompanied by an increase in throughput and reduction in scientist time required to generate the data (Fig 1B). Critically, data quality was enhanced through improved automation and application of data analysis rules and guidelines. The primary assay transitioned from a binding to a cellular assay, providing up-front human, mouse and off-target functional data for the project. These principles are being applied more widely to small molecule and antibody projects and the impact of the approach will be closely monitored.
Figure 1. The impact of changes to assay format and workflows on cycle time (A) and prouctivity (B).
1. Kola I and Landis J (2010) Nature Rev Drug Disc 3, 711-716 2. Andersson S (et al 2009) Drug Disc Today 14, 599-604 3. Sewing et al (2008) Drug Disc Today 13, 227-233
|