| 069P London, UK Pharmacology 2016 |
Molecular dynamics simulations of the μ-opioid receptor reveal distinct binding poses of structurally similar ligands
Introduction: Opioids bind to the μ-opioid receptor (MOPr), a GPCR which couples to the Gi subtype of G protein and arrestin proteins. At present the receptor conformational changes that occur following agonist binding and activation are poorly understood, particularly in relation to full versus partial agonists, or potentially biased agonists. In this study we employed molecular dynamics (MD) simulations to investigate the binding mode of two structurally similar MOPr ligands, buprenorphine and norbuprenorphine, that exhibit different efficacies for signalling outputs, in order to explore the underlying molecular mechanism.
Method: Ligand-induced Gi protein activation and recruitment of arrestin-3 was determined in vitro by a bioluminescence resonance energy transfer (BRET) assay in HEK293 cells expressing MOPr-YFP and arrestin3-Rluc, or MOPr-HA, Gi-RlucII and Gβγ-GFP. MD simulations in silicowere conducted using the antagonist-bound crystal structure of MOPr1 in the presence of ligand as the starting point. Ligands were docked using the crystallised ligand β-FNA as a template. Receptors were embedded in a POPC:POPE:cholesterol bilayer, solvated in TIP3P water and 0.15 M NaCl, and accelerated MD simulations2 were run with the Amber ff14SB forcefield3 for a total of 1 μs.
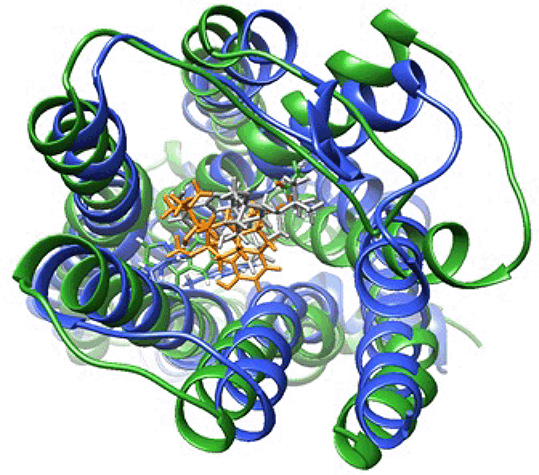
Results: The BRET assay showed norbuprenorphine is a full agonist for both Gi protein activation and arrestin-3 recruitment. Whereas buprenorphine is a partial agonist for Gi protein activation, and does not measurably recruit arrestin-3. MD simulations show that despite sharing a common morphinan scaffold, buprenorphine and norbuprenorphine adopt distinct binding poses in the ligand binding pocket (figure 1). Whilst sharing a series of common ligand-receptor interactions (e.g. D147, H297), they also interact with distinct subsets of residues. For example, norbuprenorphine but not buprenorphine interacts with N150, a key residue in maintaining the allosteric Na+-binding pocket and hence important for receptor activation4. Principle component analysis on the movements of the transmembrane helices show the buprenorphine-bound receptor occupying a distinct array of conformations from that of the norbuprenorphine-bound receptor.
Conclusion: Despite sharing a similar chemical structure, MD simulations suggest that the MOPr partial agonist buprenorphine and the full agonist norbuprenorphine occupy different binding poses and cause the receptor to explore distinct conformations during the simulation. Differences in ligand-residue interactions may underlie the differing efficacies for cellular signalling outputs for these two ligands.
References: 1. Manglik A. et al. (2012). Nature 485:321-326. 2. Pierce LCT. et al. (2012). JCTC 8:2997-3002. 3. Maier JA. et al. (2015). JCTC 11:3696-3713. 4. Fenalti G. et al. (2014) Nature 506:191-196.