Print version
Search Pub Med
| 090P London, UK Pharmacology 2017 |
A clone's genomic stability as biomarker of its DNA-damage resilience
Introduction: Dose-finding strategies in oncology seek the maximum tolerable dose assuming that “more-is-better” and “one-size-fits-all”. The height/weight of a patient are two of few individualized measures that guide dosing, with tumor-centric characteristics typically having no influence. We (1) and others have found that there is a threshold to the amount of copy number variations (CNVs) a tumor can tolerate (further referred to as genomic instability limit or GI-limit). A population’s proximity to the GI-limit has the potential to predict response to any DNA-damaging agent, for as long as CNVs are side effects of repairing therapy induced DNA-damage.
Method: We present a mechanistic pharmacodynamic (PD) model of the growth rate of a genetically homogeneous cancer cell population undergoing treatment with DNA-damaging agents. The model is formulated as a system of differential equations that link dosing to doubling time of the population. In treated populations, doubling time is decreased by a factor proportional to the likelihood that therapy will push members of the population above the GI-limit. This in turn depends on the proximity of the population to the GI-limit prior to treatment, on the cell population’s DNA-repair efficacy and on the potency of the drug-dose to increase a cell’s CNV-burden. To measure model parameters we sequence the transcriptomes of 1,569 single cells derived from human gastric cancer cell line HGC-27. This allows us to quantify CNV-burden per subpopulation and to identify cell surface markers for sorting.
Results: Clustering HGC-27 cells based on their CNV-profiles identified four groups (Fig. 1). Our PD model predicts that in the presence of DNA-damaging agents, clone 4 (n=46 cells) will grow faster than clone 1 (n=543) because of its higher DNA-repair activity (Anova:P=2E-6;F=9.78) and distance from the GI-limit (Fig. 1).
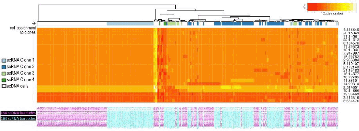
Figure 1: Clonal composition of HGC-27. CNV-landscape (rows: 22 segments) of 164 RNA-sequenced cells (columns: cyan) points to the existence of four clones (color-bars on top). DNA sequencing of 164 cells (columns: magenta) confirm the existence of the four clones.
To test this prediction we identified 24 cell surface markers that can discriminate clones 1 and 4 from the remaining clones. These markers will inform flow cytometry sorting of clone 1 and clone 4 members.
Conclusions: Our integrative systems pharmacology framework can help shed light on the relevance of pre-treatment CNV-burden on DNA-damage sensitivity.
References:
1. Andor, N., Graham, T.A., et al. (2016). Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22: 105-113.

