| 166P London, UK Pharmacology 2017 |
An analysis of the indacaterol-regulated transcriptome in human airway epithelial cells implicates gene expression changes in the beneficial and adverse effects of β2-adrenoceptor agonists in asthma
Introduction: The contribution of gene expression changes to the adverse and therapeutic effects of β2-adrenoceptor agonists in asthma is in debate1-4. Herein, we investigated if this could represent a novel mechanism of action using human airway epithelial cells as a therapeutically-relevant target.
Methods: For gene expression profiling, BEAS-2B cells were cultured for 1, 2, 6 and 18h with vehicle or indacaterol (10nM; N = 4 at each time point, ∼EC90). Total RNA was extracted and processed using Affymetrix U133plus2.0 human GeneChips. The relative expression patterning of probe sets was implemented in Transcriptome Analysis Console (Affymetrix). Functional classification of induced and repressed genes including associated gene ontology (GO) terms was performed with the database for visualization and integrated discovery bioinformatics resource. Twenty seven indacaterol-regulated genes were randomly-selected from the microarray and validated in primary airway epithelia by the low efficacy, long-acting β2-adrenoceptor agonist (LABA), salmeterol (100nM, 5 donors, ∼EC90).
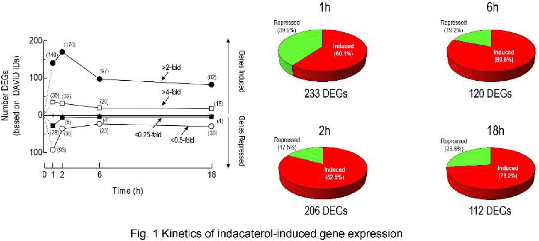
Results: Classical pharmacodynamics analyses established that the LABAs indacaterol, salmeterol, formoterol and picumeterol were full agonists on cAMP-response element reporter cells (intrinsic activity values = 1), but differed in operational efficacy5 (indacaterol ≥ formoterol > salmeterol ≥ picumeterol) by a factor of ∼10-fold. The transcriptomic signature of indacaterol identified 233, 206, 120 and 112 differentially-expressed genes (DEGs; >2-/<0.5-fold) after 1, 2, 6 and 18h of exposure respectively. Many up-regulated genes (e.g. AREG, BDNF, CCL20, EGF, EGFR, EDN1, IL6, IL9, IL20) encode proteins with pro-inflammatory activity. The enriched GO terms “signalling” and “secreted proteins” were also associated with the indacaterol-induced gene list and many of these including CRISPLD2, DMBT1, GAS1 and SOCS3 have putative in anti-bacterial anti-viral, and/or anti-inflammatory activity. Twenty seven indacaterol-regulated genes from the microarray were also found to be up- or down-regulated in primary airway epithelia by salmeterol, indicating that a genomic effect of β2-adrenoceptor agonists was a class effect and not peculiar to BEAS-2B cells.
Conclusion: Collectively, these data suggest that the consequences of inhaling a β2-adrenoceptor agonist are complex and involve widespread changes in gene expression. We propose that this represents a previously unappreciated mechanism that may contribute to the adverse-effects and therapeutic activity of β2-adrenoceptor agonists in asthma.
References:
1. Nguyen LP et al, (2017) Proc Natl Acad Sci U S A;114(43):E9163-E9171
2. Cockcroft DW, Sears MR, (2013) Lancet Respir Med;1(4):339-46.
3. Wraight JM et al, (2004) Respirology;9(2):215-21
4. Lommatzsch M et al. (2009) Thorax;64(9):763-9
5. Black and Leff, (1983) Proc R Soc Lond B Biol Sci.; 220 (1219):141-62