Print version
Search Pub Med
- M. Tsakiroglou1
- A. Doce-Carracedo1
- M. Little1
- R. Hornby1
- M. Castellanos2
- S. May2
- E. Zhang1
- K. Bloch1
- F. Miyajima1
- M. Pirmohamed1. 1Wolfson Centre for Personalised Medicine
- University of Liverpool
- Liverpool
- United Kingdom
- 2Nottingham Arabidopsis Stock Centre
- University of Nottingham
- Loughborough
- United Kingdom
| 209P London, UK Pharmacology 2017 |
Peripheral blood gene expression profiles in patients hospitalised with Clostridium difficile infection
Introduction: Clostridium difficile infection is a common adverse drug reaction caused by the use of broad spectrum antibiotics (1, 2). Differences in gene expression patterns in blood can assist in delineating mechanisms of disease pathogenesis and guide biomarker development. Transcriptional profiling of inflammatory bowel diseases has been previously explored (3-5), but diarrhoea related to C. difficile infection has been inadequately characterised.
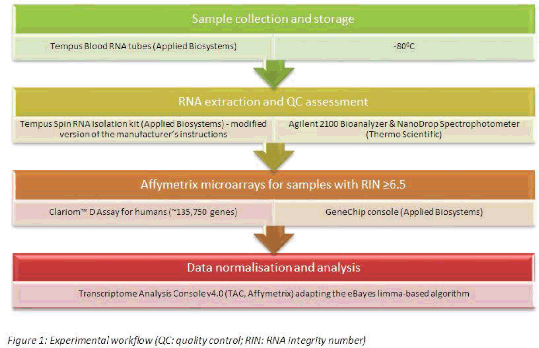
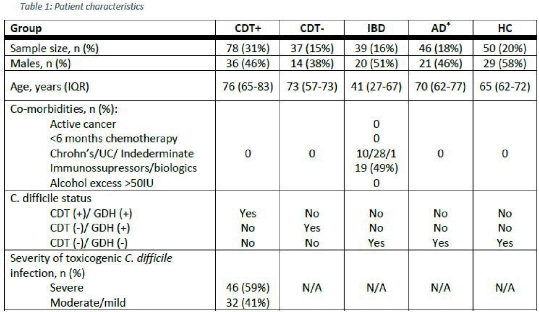
Method: Whole-blood samples were collected from patients with diarrhoea and healthy controls (HC, n=50). To achieve good representation of all-cause diarrhoea, we defined the following case populations: 1) infection with toxicogenic C. difficile (CDT+, n=78), 2) infection with non-toxicogenic C. difficile (CDT-, n=37), 3) active inflammatory bowel disease requiring hospital admission (IBD, n=39) and 4) any other cause diarrhoea from which 61% received antibiotics prior to sampling (AD, n=46). Patient characteristics and exclusion criteria are shown in Table 1. Total RNA was analysed using microarrays (Figure 1) and differentially expressed genes (DEG) were identified after applying a stringent false discovery rate (FDR) threshold of 0.01 and an absolute fold-change (FC) above 2.
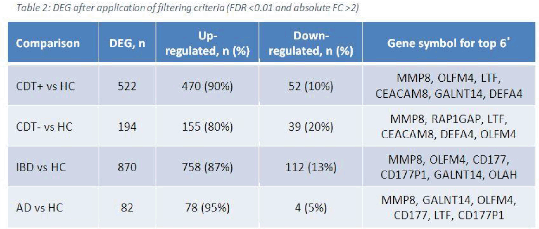
Results: Each case population was compared to the reference group of HC. Patients with IBD demonstrated the largest number of DEG followed by those with toxin-producing C. difficile infection (Table 2). The top 6 genes with the highest FC from each diarrhoea group formed a 10-gene set presented in Figure 2. Venn diagrams were utilised to identify 62 genes common to all diarrhoea groups (Table 3).
Conclusions: The majority of DEG in the IBD group compared to HC were similar to those recently published (3). Our study shows that expression is also significantly altered in patients with C. difficile infection and other causes of diarrhoea in a similar pattern. Further work is needed to determine how this relates to the pathogenesis of infectious colitis, and whether it can be used to stratify patients with different illness severities.
References:
1. Bauer MP et al. (2011). Lancet 377(9759):63-73.
2. Lessa FC et al. (2015). N Engl J Med 372(9):825-34.
3. Planell N et al. (2017). J Crohns Colitis 11(11):1335-1346.
4. Sipos F et al. (2011). Dis Markers 30(1):1-17.
5. Burczynski ME et al. (2006). J Mol Diagn 8(1):51-61.