Print version
Search Pub Med
| 024P Nottingham, UK 7th Focused Meeting on Cell Signalling |
Applying a ternary complex model to the pheromone pathway in yeast
Introduction: G-protein coupled receptors (GPCRs) remain to be a class of attractive membrane proteins for the use of whole-cell biosensors due to their responsiveness against a diverse range of ligands. Although examples are beginning to emerge in the field of synthetic biology, the pheromone response pathway in yeast remain to be underutilized for the creation of novel biosensors, despite being easily modified compared to other complex eukaryotes. Mathematical modelling can be a powerful tool to predict, clarify and provide rigorous understanding of specific relationships. An approach to utilize a combination of mathematical models and GPCR-based biosensors of yeast to demonstrate a mechanistic understanding of a complex interaction involving multiple dynamic components. Here we have modified the Bridge et al. multiple ternary complex model1 based on a set of Ordinary Differential Equations (ODEs) to recapitulate the yeast pheromone pathway.
Method: Although Bridge’s model was originally designed to demonstrate biased agonism through a dynamic multiple cubic ternary model, the model is redesigned to demonstrate the yeast pheromone pathway though a single cubic ternary model. This allows the model to demonstrate multiple states of the receptor and G-protein to understand the effects of different concentrations of each component. The GPA1 promoter library consists of creating a response curve of free Gβγ;;; concentration created by end-point time course data with an array of initial GPA1 concentrations. Computationally guided rules were then applied to experimental data created from real world biosensors in engineered yeast.
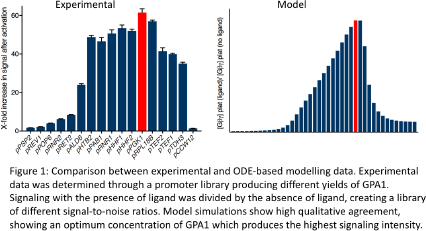
Along with the modification of reducing the model to a single compartment, a fourth pathway was added due to the Yeast having shown constitutive signaling without GPA1, which represents the capability of free Gβγ;;; being able to bind to effector proteins further down the signaling pathway. Results show similar qualitative trends to the experimental figures (Figure 1). We were able to confirm experimental behaviour against the model guided pathway which vastly improved the characteristics of the biosensor.
Conclusions: The versatility of Bridge’s model has allowed us to develop a novel type of GPCR model for yeast cells. Although the model has not fully investigated the whole signaling cascade of yeast such that in Kofal and Klipp2, focusing only on GPCR signaling and allowing multiple possibilities of constitutive activity allowed us to qualitatively describe the underlying characteristics and capabilities of yeast pheromone signaling.
References:
[1] Bridge et al. (2018) J. Theo. Biol. 442: 44-65
[2] Kofal, Klipp. (2004) Yeast 21: 831-850

