| 003P London, UK Current trends in Drug Discovery – Young Scientists and Tomorrow’s Medicines |
Characterising the catalytic mechanism of a Methyltransferase for use in oncology drug discovery
Introduction: Understanding the catalytic mechanism of target enzymes in drug discovery enables development of balanced assays for hit finding screens and aids profiling mechanisms of new inhibitors, so could improve translation to the clinic.1 Mechanisms of bi-substrate bi-product enzymes include ternary complex formation, either by random or sequential binding, or a Ping Pong mechanism where two half reactions occur with a covalently bound intermediate. The aim of this study was to elucidate the mechanism of a Methyltransferase; an attractive enzyme class of epigenetic oncology targets.2
Methods: Using a discontinuous MTase-Glo™ (Promega) assay3, human MT was studied with two substrates: S-Adenoysl Methionine (SAM), and a 21 residue histone peptide with a methyl-accepting residue. Initial rates were examined across a range of substrate concentration combinations and data fitted to ternary Bi Bi and Ping Pong models (Prism), and compared using Akaike’s Information Criteria. SAH and the methylated peptide were used as product inhibitors. Dead-end analogues were 5'-methylthioadenosine and a mutant peptide with the methyl-accepting residue replaced with lysine. Initial rate data were globally fit to competitive, non-competitive, mixed and uncompetitive inhibition models and compared using an F test.
Results: Initial velocity data fit best to the Ping Pong model (Figure 1) suggesting formation of a methyl-bound MT intermediate. Product inhibition was consistent with the Theorell-Chance, Ping Pong, and Rapid Equilibrium random mechanism with dead-end EAP and EBQ complexes (Table 1). Lack of uncompetitive inhibition by dead-end inhibitors indicated random order substrate binding, ruling out Ping Pong and Theorell Chance mechanisms. Pre-steady state kinetics revealed a burst phase, indicating a rate limiting step other than catalysis.
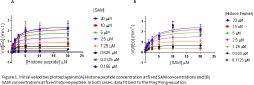
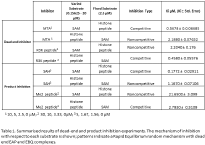
Conclusion: Multiple approaches are needed to elucidate the catalytic mechanism of an enzyme. Inhibitor studies suggest this MT uses a Rapid Equilibrium Random mechanism with dead-end EAP and EBQ complexes.
References:
1. Holgate, G et al. (2017) Nature Reviews Drug Discovery 17: 115-132
2. Blanc, R et al. (2017) Molecular Cell 65: 8-24
3. Hsiao, K et al. (2016) Epigenomics 8: 321-339

