| 1177 Virtual Meeting BPS & ELRIG UK joint meeting: Translating Ideas into Therapies |
Development of Robust and Predictive Machine Learning QSAR Models for Hepatic Stability
Introduction/Background & aims Assessment of pharmacokinetic properties of compounds is a critical step in drug discovery. Measuring hepatic stability is essential in establishing the permanence and clearance of a drug in the body. Usually, this endpoint is evaluated in vivo, using rats, or in vitro, using human liver microsomes. Recently, in silico approaches have earned prestige in evaluating the pharmacokinetic properties of bioactive compounds [1]. Herein, we describe (i) the collection, curation, and integration of the largest publicly available dataset of human hepatic stability measured in vitro with liver microsomes and (ii) the development and statistical validation of robust and predictive QSAR models for this endpoint.
Method/Summary of work Human liver microsome (half-life, T1/2) data was compiled from ChEMBL (IDs CHEMBL2367379 and CHEMBL613373), totaling 8,023 datapoints. The data was curated following a protocol developed by our group. During this process, we evaluated the concordance of assays (n=4,511), eliminated inconsistent data (n=2,650), removed mixtures, inorganics, and counter ions, and normalized specific chemotypes (n=2,639), and removed duplicates (n=2,432). Compounds were classified as stable if T1/2 values > 30 min, resulting in 37% classified unstable and 63% classified stable compounds. QSAR models were developed in KNIME employing three types of molecular descriptors (Morgan and MACCS fingerprints, and RDKit properties) using Random Forest as the machine learning algorithm. Models were developed and validated following the best practices proposed by OECD [2] and by employing 5-fold external cross-validation procedure to estimate the robustness of the developed models. Moreover, we estimated the applicability domain (AD) [3] and performed 20 rounds of Y-randomization to ensure the absence of chance correlations.
Results/Discussion The statistical characteristics of the models are shown in Fig. 1. All models were robust and predictive. Morgan fingerprints showed the highest correct classification rate (CCR) of 0.78, sensitivity of 0.89, specificity of 0.67, coverage of 1.0, and positive and negative predictive values (PPV and NPV) of 0.82 and 0.78, respectively.
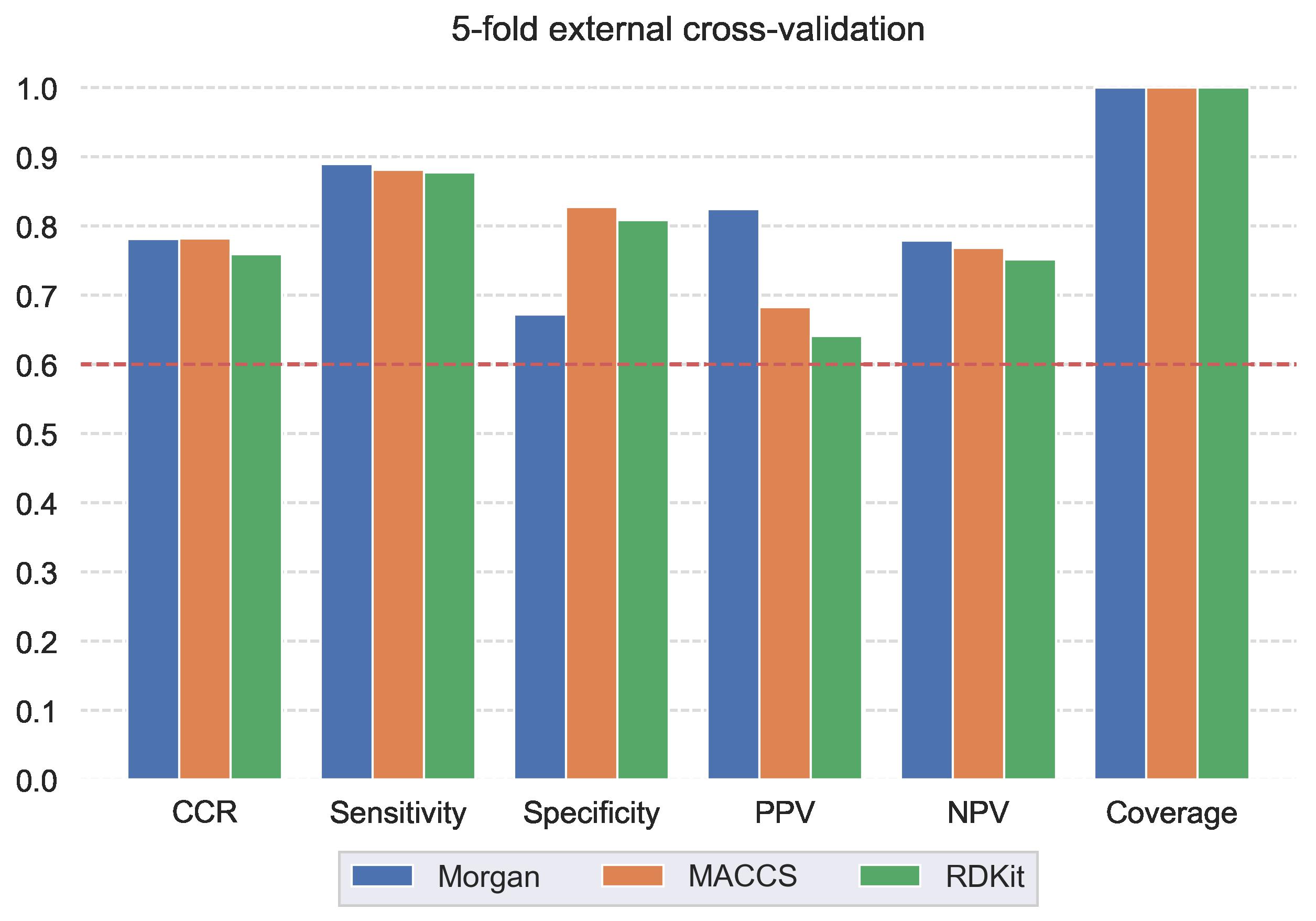
Figure 1. Statistical characteristics of the QSAR models developed for hepatic stability.
Conclusion(s) We collected, curated, and integrated the largest publicly available dataset for human hepatic stability and developed robust and predictive QSAR models. These can be employed to predict the half-life of compounds in human liver microsomes with high accuracy. The models will be implemented as part of a comprehensive platform for the prediction of pharmacokinetic parameters, which will be freely available for the scientific community.
Reference(s)
1. Liu, R.; Schyman, P.; Wallqvist, A. Critically Assessing the Predictive Power of QSAR Models for Human Liver Microsomal Stability. J. Chem. Inf. Model. 2015, 55, 1566–1575, doi:10.1021/acs.jcim.5b00255.
2. Tropsha, A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010, 29, 476–488, doi:10.1002/minf.201000061.
3. Tropsha, A.; Golbraikh, A. Predictive QSAR modeling workflow, model applicability domains, and virtual screening. Curr. Pharm. Des. 2007, 13, 3494–504.

