Print version
Search Pub Med
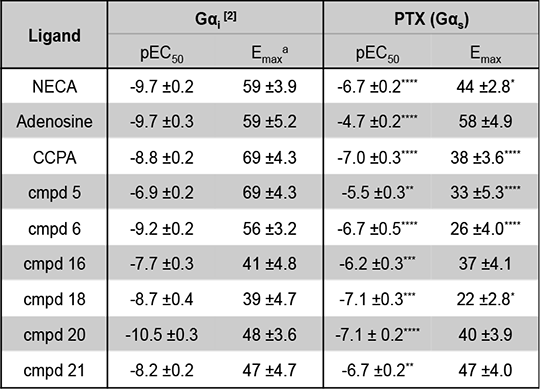
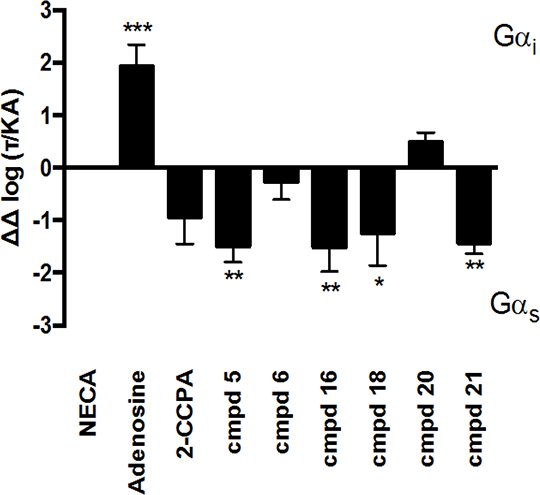
Characterising signaling bias of novel Adenosine A1 receptor agonists The adenosine receptors (ARs) are family A G protein-coupled receptors (GPCRs) that exist as 4 subtypes; A1, A2A, A2B and A3, each with ~50-60% homology [1]. All four ARs modulate cAMP levels, with the A1R and A3R coupling to Gαi reducing cellular cAMP, while the A2Rs stimulate cAMP production via Gαs activation. All ARs are activated by the purinergic nucleotide, adenosine, and 2 synthetic adenosine derivatives; 5’-N-ethylcarboxamidoadenosine (NECA) and chloro-N6 -cyclopentyladenosine (CCPA). We have developed novel AR ligands, some of which display A1R selectivity [2]. In an attempt to investigate which Gαi/o proteins couple to the A1R we used pertussis toxin (PTX), to inhibit the action of these, at the A1R. CHO cells, stably expressing the A1R (CHO-A1R), treated in this manner, were subsequently stimulated with our ligands, and cAMP levels measured. Interestingly we observed cAMP production upon PTX treatment (Table 1.), indicative of a Gαs component to the A1Rs repertoire. Whilst this has been observed before [3] it has not been seen to such significant levels, and has not been quantified. Thus here we present a quantification of the signaling bias between the canonical, Gαi, pathway, and non-canonical Gαs (Figure 1.). It is interesting to note that the ligand that displays the most extensive Gαi is the natural cognate ligand, adenosine. These agonists are currently the focus of on-going studies pertaining to; calcium mobilisation, ERK activation, and which inhibitory G proteins they activate.
Table 1. Potency and Emax for CHO-A1R cells a - Emax for Gαi coupling calculated as percentage inhibition when stimulated with 10μM forskolin.
Figure 1. Relative bias for the A1R. Signalling bias was calculated relative to NECA as ΔΔ log(τ/KA) [4]. Statistical difference to NECA was determined using a one-way ANOVA with a Bonferroni’s post-test (*, p < 0.05, **, p < 0.01, ***, p ,0.001). Data are the mean of 5 individual data sets ± SEM.
[1] Jacobson and Gao (2006) Nat Rev Drug Discov 5:247–264, [2] Knight et al (2016) J Med Chem 59:947-964, [3] Baker and Hill (2007) J Pharmacol Exp Ther 320:218–282, [4] Baltos et al (2016) Biochem Pharmacol 99:101–112. Work funded by the BBSRC (G.L - BB/M00015X/1) and MRC (I.W - MR/J003964/1).
|